And a global consortium of industry and academic partners

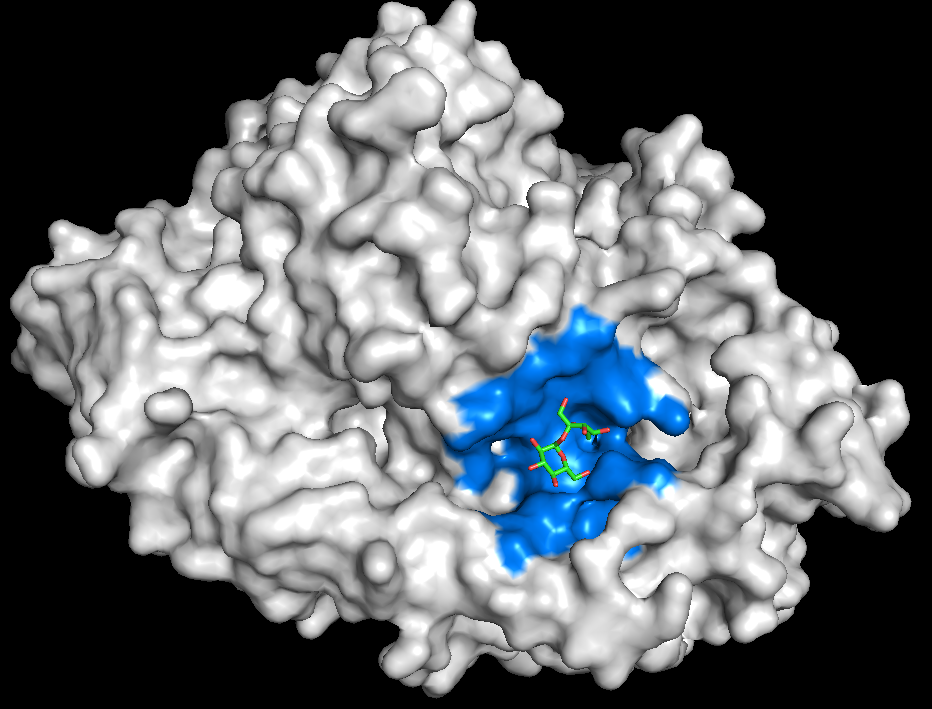
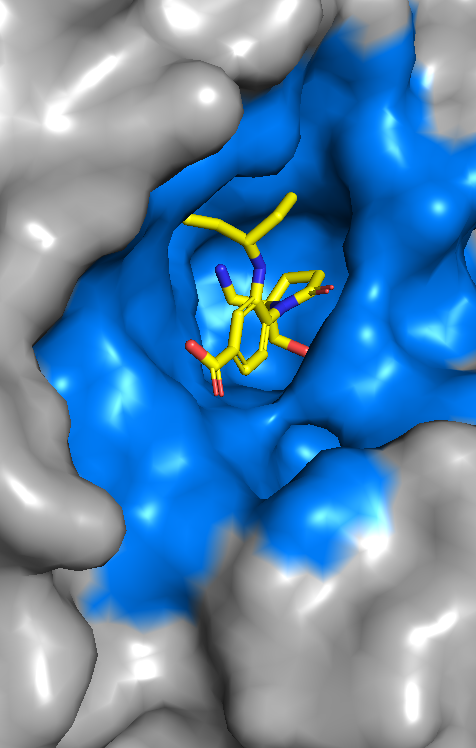
AQAffinity is an open-source AI model for fast, accurate, prediction of drug binding affinities without experimental structures. Built on top of the OpenFold Consortium’s OpenFold3 co-folding model, AQAffinity enables scalable potency prediction for virtual screening. AQAffinity expands the functionality of the OpenFold3 model to match the state-of-the-art in structure-aware AI while remaining fully open and accessible.
By enabling rapid, binding-affinity prediction directly from sequences, AQAffinity dramatically accelerates early-stage drug discovery. It helps researchers prioritize promising candidates sooner, reduce costly wet-lab cycles, and ultimately bring effective therapies to patients faster.
Accurate binding affinity prediction is a critical bottleneck in early-stage drug discovery. Experimental screening is expensive and slow, while traditional computational methods such as free energy perturbation (FEP) are too costly to deploy at scale and require highly accurate structural input.
Recent advances in deep-learning–based structure prediction have created a new opportunity: combining predicted structural information with large-scale experimental activity data to enable practical, scalable affinity prediction.
AQAffinity brings this capability to the OpenFold3 ecosystem.
AQAffinity is now available fully open-sourced under the permissive Apache 2.0 license on Hugging Face for academic and industry researchers and developers globally.
We can’t wait to see what you will build using AQAffinity.
Learn more about AQAffinity:
with Nvidia and the OpenFold Consortium
